Environmental Molecular Sciences Laboratory
Agricultural Soils from the Southern Great Plains Are Efficient Ice Nucleating Particles

Direct measurements of ice nucleating particle composition show agricultural soils were highly efficient ice nucleators.
Natural and Cultivated Marine Algae Have Substantially Different Metabolites
Single-colony analysis reveals metabolic differences between natural and cultured algae.
Laser Capture Dissection Microscope
The Zeiss Laser Capture Dissection Microscope allows for the spatially resolved collection and enrichment of unique populations of cells from a thin section of tissue, biofilm, or monolayer of cells. Samples can be collected based on specific morphology or fluorescent labeling. Enriched populations can then be used for propagation or omics analysis.

Research application
- Supporting the Biomolecular Pathways Integrated Research Platform, this resource enables the isolation of specific cell-types or single cells for further omics analyses such as cell-specific proteomics or single-cell RNA-Seq for transcriptional analysis.
- Supporting the Cell Signaling and Communication Integrated Research Platform, this resource supports studies on the spatial organization of tissues and microbial communities to understand the structure-function relationships and the molecular pathways within and between cells.
- Supporting the Rhizosphere Function Integrated Research Platform, this resource provides new insights to the interactions between plant tissue and the microbial community, such as interactions between roots and nitrogen-fixing bacteria, to better understand the molecular mechanisms underlying plant productivity and resilience to environmental stress.
Available Instruments
Zeiss Laser Capture Dissection Microscope
Proposal Guidance for EMSL-JGI FICUS Program
Discovery of a Novel and Sustainable Organic Liquid Hydrogen Carrier

Scientists suggest a new way of binding hydrogen with compounds by using lignin-based jet fuel that enables stable storage and transportation of hydrogen.
X-ray Photoelectron Spectroscopy

X-ray photoelectron spectroscopy (XPS) is a widely used surface analysis technique providing information on the chemical nature and state of detected elements. It is a useful method to determine the surface composition of bio-organic systems and determine organic compositions on mineral surfaces. XPS is available to the research community at the Environmental Molecular Sciences Laboratory (EMSL) through open calls for proposals.
With the use of XPS, chemical information obtained can be further upgraded by expressing the XPS data in terms of concentrations of ingredients, classes of biochemical compounds, or various organic and inorganic compounds.
Imaging XPS system provides fast, quantitative, real-time imaging with high spatial resolution. It can quantitatively determine a sample’s elemental composition within the first 10nm of a surface and allows high-resolution scans to be taken of elemental peaks of interest, determining valuable chemical bonding information.
Research application
XPS can be used to study the biogeochemical and microbial processes that drive fate and transformations of nutrients in soil and shallow subsurface environment. This technique can contribute to understanding the mechanisms and biogeochemical conditions by which organic matter and microbial residues are stabilized or released by mineral surfaces and interfaces. This technique can also be used to study the surface composition of microorganisms to better understand a wide range of interactions involving the cells (aggregation, adhesion, biofilm formation, and infection) and the relationships between surface composition and surface structure.
- XPS supports the Terrestrial-Atmosphere Processes Integrated Research Platform by analyzing a wide range of chemical and spatial information—helping to uncover and further understand the drivers of various environmental fluxes, including atmospheric and soil interactions among others.
- XPS supports the Biogeochemical Transformations Integrated Research Platform by enabling new insights into molecular structure and activity, while also helping to answer fundamental questions about environmental chemical, physical, hydrologic, microbial, and atmospheric interactions. Additionally, EMSL offers a sample preparation workflow that allow researchers to rapidly analyze (in high throughput manner) the chemistry of soil organic matter associated with mineral surfaces.
Available instruments
- Temperature Programmed Desorption
- Ultraviolet Photoelectron Spectroscopy
- Icon Scattering Spectroscopy
- Fast entry load lock with pre-cooling device and full-size glove box
- Cluster/Ar+ ion gun
- Monchromated X-Ray Gun (Al and Ag anodes)
Tips for success
- Sample preparation, history, and providence must be discussed prior to sample submission and user analysis. Contact your science point of contact for sample-specific requirements.
Contributing team and resources
EMSL develops and deploys capabilities for the user program by conducting original research independently or in partnership with others and by adapting/advancing science and technologies developed outside of EMSL. In some instances, EMSL directly deploys mature capabilities developed by others where there is value for the EMSL user community. The following grants/activities, PI’s and teams contributed to the development of this capability:
- Qian Zhao and Mark Engelhard, DE-AC05-76RL01830, Environmental Molecular Sciences Laboratory
- Qian Zhao and Arunima Bhattacharjee, DE-AC05-76RL01830, Environmental Molecular Sciences Laboratory
Related publication
Cheng, C. H., Lehmann, J., & Engelhard, M. H. (2008). Natural oxidation of black carbon in soils: changes in molecular form and surface charge along a climosequence. Geochimica et Cosmochimica Acta, 72(6), 1598-1610. https://doi.org/10.1016/j.gca.2008.01.010
Data Visualization
EMSL pushes the boundaries of scientific discovery by creating a broad range of visualizations for experimental results—including modeling of molecular interactions, ab initio simulation, and visual and statistical integration of disparate data types. EMSL has developed software packages for advanced data visualization, and computational and data scientists are available to provide expertise and guidance. EMSL data visualization resources complement the standard data processing, analysis, and visualization that is typically performed by instrument scientists as part of sample analysis.
Research application
The Data Transformations Integrated Research Platform supports EMSL’s visualization resources, which are vital in advanced data analytics, simulation, and integration.
These tools support our expertise across all Integrated Research Platforms, but are especially useful in
- Biomolecular Pathways Integrated Research Platform: Robust statistical analysis and visualization of organic matter function and composition.
- Rhizosphere Function Integrated Research Platform: Visualizations for spatially resolved genotypic and phenotypic traits.
- Biogeochemical Transformations Integrated Research Platform: Computational chemistry tools and models across scales.
Available Resources
Tips for success
To best use these visualization capabilities, it is recommended you speak with the integrated research platform lead for specific guidelines applicable to your project.
- Reach out to your EMSL science point of contact at the beginning of your project. By engaging early, data can be analyzed as expeditiously as possible.
- Visualization, analysis, and collaboration throughout the life of the project will help guide ultimate analysis.
Contributing Teams and Resources
EMSL develops and deploys capabilities for the user program by conducting original research independently or in partnership with others and by adapting/advancing science and technologies developed outside of EMSL. In some instances, EMSL directly deploys mature capabilities developed by others where there is value for the EMSL user community. The following grants/activities, PI’s and teams contributed to the development of this capability:
- Ryan Hafen, https://trelliscope.org/
- David Degnan, Environmental Molecular Sciences Laboratory, https://shinyproxy.emsl.pnnl.gov/app/pspecter
- Ljiljana Paša-Tolić (HuBMAP PI), David Degnan (developer), https://github.com/PNNL-HubMAP-Proteoform-Suite/IsoMatchMS
- Lisa Bramer, Project 51146, Environmental Molecular Sciences Laboratory and Microbes in Transition LDRD (90001 – N56672) https://shinyproxy.emsl.pnnl.gov/app/freda
- Lisa Bramer (PI) and David Degnan (developer), Project 51146 Environmental and Molecular Sciences Laboratory, https://map.emsl.pnnl.gov/app/mode-classic (GUI for Trelliscope displays, under development/updates)
Related Publications
Hafen, Ryan, et al. "Trelliscope: A system for detailed visualization in the deep analysis of large complex data." 2013 IEEE Symposium on Large-Scale Data Analysis and Visualization (LDAV). IEEE, 2013. DOI: 10.1109/LDAV.2013.6675164.
Degnan, David J., et al. "PSpecteR: A User-Friendly and Interactive Application for Visualizing Top-Down and Bottom-Up Proteomics Data in R." Journal of Proteome Research 20.4 (2021): 2014-2020.
Degnan, David J., et al. "IsoMatchMS: Open-Source Software for Automated Annotation and Visualization of High Resolution MALDI-MS Spectra." Journal of the American Society for Mass Spectrometry 34.9 (2023): 2061-2064.
Bramer, Lisa M., et al. "ftmsRanalysis: An R package for exploratory data analysis and interactive visualization of FT-MS data." PLoS computational biology 16.3 (2020): e1007654.
Molecular Observation Network (MONet)
Environmental Transformations and Interactions

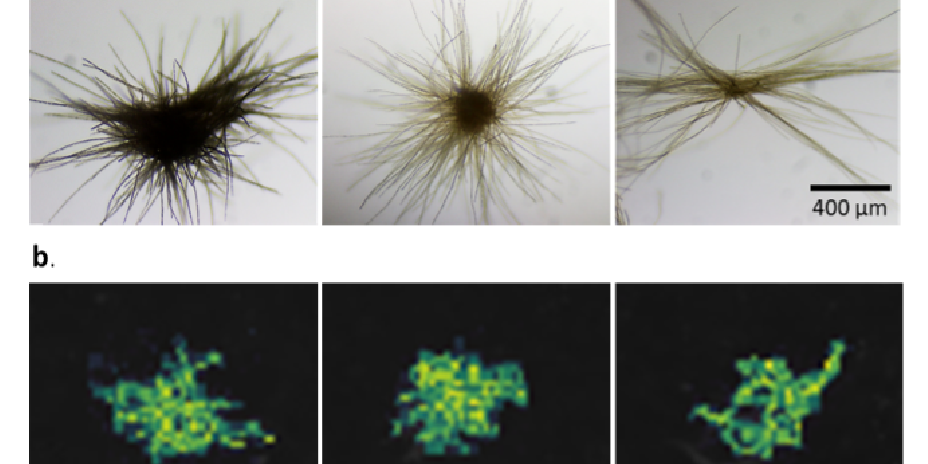
New Workflow for X-ray Photoelectron Spectroscopy Allows for Soil Analysis

Sample preparation method allows for analysis of up to 50 soil samples per day



